Catch-up On xMAP® Connect 2025: Eight Virtual Presentations to Share Insights
By Marcha van der Steen
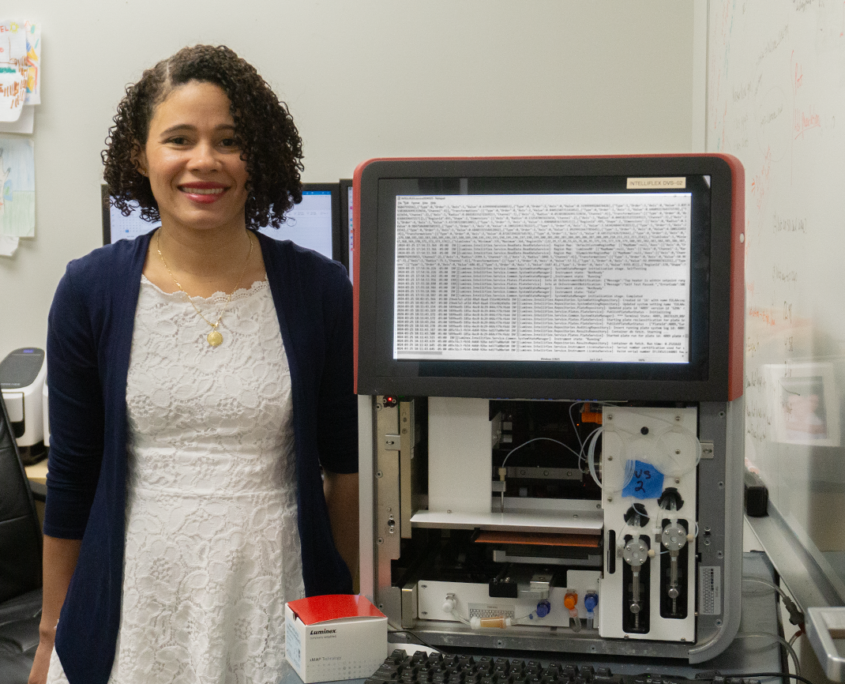
Sherry Dunbar kicked off the series with a talk about the history of xMAP® Technology and some of the latest published studies that cite the technology

The community of researchers using xMAP® Technology to advance their work is truly impressive and productive! Since the technology was first invented in the 1990s, it has been cited in more than 105,000 peer-reviewed studies, with a worldwide installed base of more than 20,000 instruments. It comes as no surprise that great things happen when the xMAP community gets together to share expertise and exchange ideas.
xMAP® Connect Virtual 2025
For 2025 xMAP Connect, we took a new approach. We replaced our traditional regionalized in-person events with an 8-week virtual series, releasing a new presentation online each Wednesday throughout October and November. All sessions are now available on-demand. We hope this new format serves our community well and provide valuable insights for potential new users.
A deep dive into xMAP Technology with Sherry Dunbar
The first presentation came from xMAP aficionado Sherry Dunbar, Senior Director for Luminex’s scientific affairs programs. She has been with the company since 1999, when the very first xMAP reader was launched. In her talk, she walks quickly through the history of xMAP instrumentation — from that first Luminex 100 platform to the latest generation, the xMAP INTELLIFLEX® System — and consumables. If you’ve ever wondered exactly how this technology works, this is the presentation for you! From the liquid suspension format to laser analysis to dual reporter channels, Dunbar’s talk has you covered.
The presentation also compares xMAP Technology to other approaches. Compared to single-plex tests, xMAP assays use less sample input, require less time and labor, and reduce cost, all while producing more data with greater flexibility and improved reproducibility. When compared to planar arrays, xMAP offers a liquid-phase suspension that delivers faster kinetics and higher surface-to-volume ratio.
One of the reasons that xMAP has been so widely adopted is its remarkable versatility. It can interrogate proteins or nucleic acids, making it a great fit for proteomic applications, such as studies of cytokines or immunoglobulins, as well as genomic applications, including gene expression and SNP genotyping, among others.
Data is in the details
Dunbar’s presentations often kick off our xMAP Connect events, and her journal club roundup of some recently published studies is always a crowd favorite. This year, she focused on three areas where use of xMAP Technology is growing rapidly: biopharma, vaccine research and development, and diagnostic laboratories. Here are some of the highlights.
Scientists from Arbutus Biopharma offer preclinical characterization of an siRNA therapeutic candidate that has been developed to address chronic cases of hepatitis B. An analysis of cytokines demonstrated a lack of off-target effects, indicating that the siRNA therapy does not trigger an unwanted immune reaction.
At Moderna, researchers developed an mRNA vaccine against RSV, which is especially dangerous for children and the elderly. The paper reports robust data from rodent models, which led to clinical evaluation of the RSV vaccine candidate in adults and children. In a phase 2/3 trial, the vaccine proved effective in protecting against RSV in people aged 60 or older.
Researchers at Case Western Reserve University School of Medicine designed a study to assess the societal impact of grants funded through the Clinical and Translational Science Award (CTSA) program, using a local laboratory as a microcosm. They determined that the program led to better public health and local economic benefits.
This year’s virtual xMAP Connect presentations may be over however, they are now available on-demand. Watch now!
Can’t get enough of xMAP Connect? Recordings of xMAP Connect presentations from 2024 and previous years are also available for on-demand viewing.
* For Research Use Only. Not for use in diagnostic procedures.
† Luminex does not endorse any specific LDT. Performance characteristics of Laboratory Developed Tests are determined by the laboratory.